


- Multiple sequence alignment manual#
- Multiple sequence alignment full#
- Multiple sequence alignment download#
Using the default parameters, ClustalW2 returns the following alignment: Make the alignment using default parameters.
Multiple sequence alignment full#
If you want to do full iterations (guide tree and MSA) choose TREE By default, ClustalW performs progressive alignment (no iterations) but you can use iterations by changing the iteration type parameter (green).You can also define a minimal distance between gaps ( gap separation distance): gaps that are closer than this distance are extra penalized except when they are at the ends of the alignment ( no end gap separation penalty). G-rich regions or hydrophilic regions are more likely to contain gaps so gap penalties are reduced in such regions. For proteins, a complex scoring system is used with specific gap penalties for regions that are rich in one (type of) amino acid e.g. To make these pairwise alignments a scoring system is used that you can define (red).
In contrast to Clustal Omega, which replaces amino acids by numbers to speed up pairwise similarity score calculations, ClustalW builds the guide tree based on real pairwise alignments. Now we will make the alignment using ClustalW2. if the amino acids on a position are different: a - is printedĪbove the consensus sequence you see the conservation scores that represent the similarity on each position: the higher the score the more similar the amino acids on that position are.if all amino acids on a position are similar: a + is printed.if the majority of the sequences contain the same amino acid on a position: the amino acid is printed in small letters.if each sequence contains the same amino acid on a position: the amino acid is printed in capitals.amino acids with aromatic side chains (F, Y and W) are coloured orangeĪt the top of the alignment you see the consensus sequence giving a one line representation of the alignment:.amino acids with polar uncharged side chains (S, T, N and Q) are coloured green.positively charged amino acids (R, H and K) are coloured blue.negatively charged amino acids (D and E) are coloured red.The Zappo scheme colors the sequences according to their biophysical properties: The sequences in this example are too short and simple to see the impact of iterations so we are going to use the default settings.Īlign these sequences with ClustalOmega using default parameters. That's why the default setting is to not use the iterations. Each iteration will add 1-3 times it costs to make the alignment without iterations. Iterations come at a cost, they increase the run time. The number of times it refines the alignment is defined by the Max number of HMM iterations parameter. Apart from refining the guide tree, Clustal Omega will also refine the MSA based on the guide tree by randomly removing one sequence from the MSA, making a (hidden Markov) model of the remaining alignment and realigning the removed sequence to the model.The number of times it performs this step to change (and improve) the guide tree is defined by the Max number of guide tree iterations parameter. Clustal Omega generates a guide tree (like all MSA algorithms) but it changes this tree by replacing the two most similar sequences by a model that represents their alignment and recalculating the guide tree but now with the model instead of with the two separate sequences.The number of iterations to improve the MSA are defined by the Number of iterations parameter.Parameters of Clustal Omega.Ĭlustal Omega has the following parameters: The tools for multiple sequence alignment can be found in the Tools menu.
Multiple sequence alignment download#
You can download the sequences in fasta format. We are first going to use different tools for the toy example below:
Multiple sequence alignment manual#
Some MSA tools allow manual editing of alignments, which will be discussed in the next exercise.
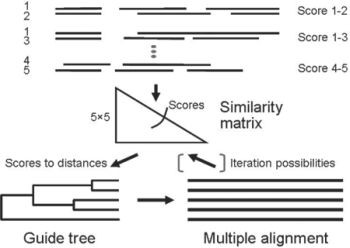
Let's discover how some of these programs behave, and which suit your needs best! As always, EBI provides a nice collection of web-based alignment tools but you can also make MSAs in Ugene. Fortunately, you can also find tools to convert MSA formats. Several multiple sequence algorithms exist, each with their own program and format.


 0 kommentar(er)
0 kommentar(er)
